qPCR Extraction Control Red
Molecular IVD companies continue to experience tighter and tighter regulations in regard to incorporating appropriate positive and negative controls into their assays.
Meridian’s qPCR Extraction Control Red uses E. coli cells of a known concentration that contain quantitative PCR positive control DNA sequence (with no known homology to any organism, so that it does not interfere with the detection of the sample target DNA). Their composition closely resembles a test sample and serves as a full-process quantitative PCR positive control, monitoring the success of a qPCR assay from lysis/extraction to amplification.
Have questions about a product?
Contact us to learn more about Meridian’s molecular or immunoassay reagent portfolio. We want to hear from you!
Workflow
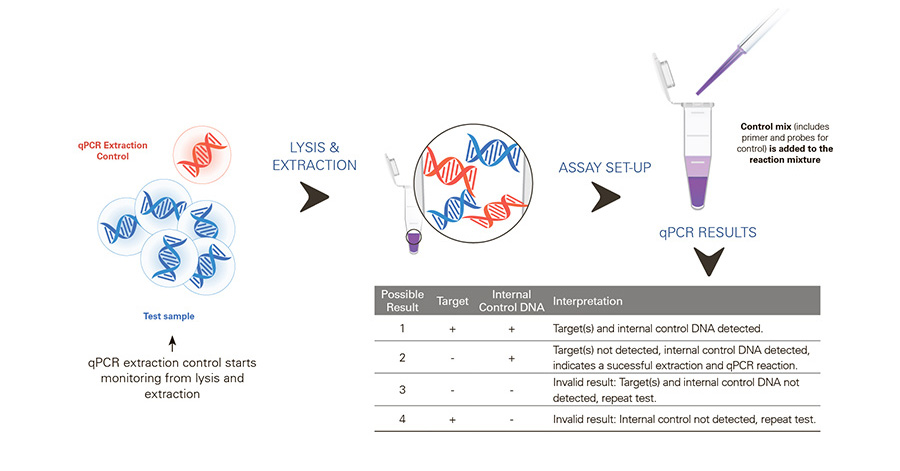
qPCR Extraction Control Red, MDX026
Control used to monitor the success of a qPCR assay from lysis/extraction to amplification and reduces the chance of obtaining false negative results from the sample DNA.
Documents & Resources
Description
qPCR Extraction Control Red comprises internal control DNA sequence in E. coli cells and a control mix (containing specific primer and probe (emission wavelength = 670nm)) for qPCR detection. The cells are added to the sample and lysed and extracted with the sample DNA for downstream amplification by qPCR. The detection of qPCR Extraction Control acts as a PCR positive control and confirms the success of the extraction and amplification steps, avoiding the misinterpretation of a false negative result.
Specifications
| Description | Control used to monitor the success of a qPCR assay from lysis/extraction to amplification and reduces the chance of obtaining false negative results from the sample DNA. |
| Concentration | 25x |
| Appearance | Clear, colorless solution |
| Internal Control DNA | In E. coli cells (genotype: F- deoR endA1 recA1 relA1 gyrA96 hsdR17(rk-, mk+) supE44 thi-1 phoA Δ(lacZYA-argF)U169Ф80lacZΔ15λ–pBR322 (ranseqb1 AmpR)). |
| Control Mix | Primers and Quasar® 670 labeled probe |
| Application | Probe-based, qPCR |
| Sample type | Tissue, cells |
| Presentation | 2 vials |
| Storage | -20 °C |
| Mix stability | See outer label |
| Functionaity/ Consistency | Ct of 29 ± 1.6. The difference between test and history is less than 1 Ct. |
Catalogs & Brochures
qPCR Extraction ControlsqPCR Extraction Controls
FAQs: qPCR Extraction Control
The benefit of the artificial qPCR Extraction Control cells in evaluation of the extraction process is the possibility to validate sample extraction process. Signal derived from the qPCR Extraction Control confirms the success of the extraction step and monitors co-purification of PCR inhibitors that may cause biased or PCR false negative results. Housekeeping genes or spike in controls do not monitor the whole extraction process, especially the lysis step and in the case of housekeeping genes, low extraction efficiency and degraded samples.
When performing DNA extraction, it is often advantageous to have an exogenous source of DNA template that is spiked into the sample prior to lysis. This control DNA is then co-purified with the sample DNA and can be detected as a PCR positive control for the whole extraction process.
Although it is not mandatory yet, regulators across the world are tightening up recommendations for the addition of internal controls, EU: IVDD to IVDR 2017/746 “Control materials with quantitative or qualitative assigned values intended for one specific analyte or multiple analytes shall be classified in the same class as the device.” MDSAP with FDA (“general controls ensuring effectiveness of the device”), TGA (“provisions for the user, at time of use, of device performance”), MHLW and ANVISA
Yes, it is possible that some of the genes will be the similar, the genotype of the E. coli that we use is F- deoR endA1 recA1 relA1 gyrA96 hsdR17(rk-, mk+) supE44 thi-1 phoA Δ(lacZYA-argF)U169 Φ80lacZΔM15 λ- pBR322 (ranseqb1 AmpR). You will need to check before using the qPCR Extraction Control with other bacteria.
The primers and probe for the qPCR Extraction Controls are in the Control Mix.
We recommend using the probe-based detection method for quantification of DNA extraction controls, but it would be possible to use other PCR applications for detection of the extraction controls, such as an intercalating dye such as SYBR® Green (but this would be singleplex), or endpoint PCR.
The two colors are for the fluorescent dyes on the qPCR Extraction Control probe. Control Mix Red (Cy5 – emission wavelength = 670nm) and Control Mix Orange (HEX – emission wavelength = 560nm). This allows you to chose one that will fit with your existing protocol in a multiplex qPCR assay.
No, the qPCR Extraction Control will work with any commercially available DNA extraction method, such as silica-membrane extraction kits, magnetic bead-based extraction, phenol-based extraction and CHELEX matrices and has been tested on a wide range of qPCR platforms.
Get In Touch With A Specialist
Have questions about a product? Want to learn more about Meridian’s molecular or immunoassay reagent portfolio? We want to hear from you!