Glycerol-Free T7 RNA Polymerase (HC)
T7 RNA Polymerase is a DNA-dependant RNA Polymerase originating from the T7 bacteriophage and is the industry standard enzyme for in vitro transcription (IVT). Not only does T7 RNA Polymerase provide for mRNA vaccine production, but it can also be used for other RNA-based therapeutic and research applications, including miRNA and siRNA synthesis for RNA interference drugs, synthesis of sgRNA for the CRISPR/Cas9 system and in isothermal diagnostic methods, such as nucleic acid sequence-based amplification (NASBA).
Have questions about a product?
Contact us to learn more about Meridian’s molecular or immunoassay reagent portfolio. We want to hear from you!
RNA Yield and Integrity
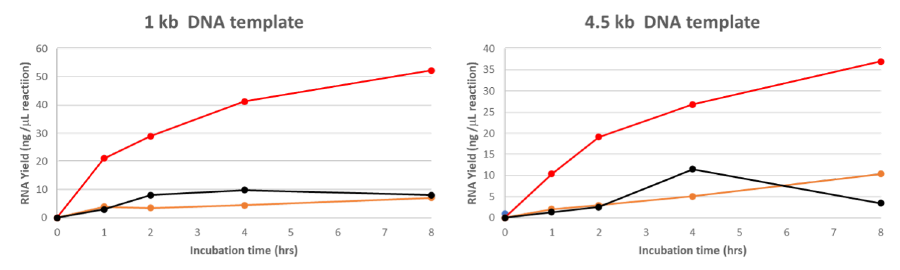
In vitro transcription reactions were assembled, according to the manufacturer’s recommended protocol, using 0.1 ng of dsDNA template encoding a 1 kb or 4.5 kb RNA. The reactions were incubated at 37°C for up to 8 hours with lyophilized Glycerol-Free T7 RNA Polymerase (HC) (MDX201) (red), NEB T7 RNA Polymerase (orange) and ThermoFisher T7 RNA Polymerase (black). These samples were tested by performing In vitro transcription reactions against MDX201 before lyophilization, NEB T7 RNA Polymerase and ThermoFisher T7 RNA Polymerase. Transcription reactions were purified using a column-based RNA purification kit and quantified on spectrophotometer. These results demonstrate that a considerably higher yield of RNA was synthetized using MDX201, and that MDX201 provide a greater transcript integrity after extended duration of RNA synthesis reactions compared to other manufacturers’ kit.
Glycerol-Free T7 RNA Polymerase (HC), MDX201
Glycerol-free, high concentration DNA-dependant RNA Polymerase that requires a T7 promoter sequence, it an ideal enzyme for in-vitro synthesis of labeled or unlabeled RNA for research, diagnostic or clinical applications. Ideal for lyophilization.
Documents & Resources
Glycerol-Free T7 RNA Polymerase (HC) is a high concentration (1,000 U/µL) DNA-dependent RNA Polymerase with very high specificity for the T7 phage double-stranded promoter sequence. The enzyme catalyzes the 5’→3’ synthesis of RNA on either single-stranded DNA or double-stranded DNA downstream from the promoter. Glycerol-Free T7 RNA Polymerase (HC) has been optimized to deliver excellent, stable performance without glycerol, allowing you to develop in vitro transcription workflows with a high-quality enzyme that delivers reliable results with the added capability for lyophilization. This gives you the flexibility to confidently store reaction mixtures at ambient temperature or produce diagnostic assays that rely on miniaturized reaction components.
Catalogs & Brochures
Glycerol-Free T7 RNA PolymeraseGlycerol-Free T7 RNA Polymerase
A Comprehensive Guide to Essential Reagents for High-Throughput Molecular Diagnostic Assays (MDx)Accelerating Efficiency: A Comprehensive Guide to Essential Reagents for High-Throughput Molecular Diagnostic Assays (MDx)
FAQs: Glycerol-Free T7 RNA Polymerase (HC)
RNA polymerase from a T7 bacteriophage is very selective and efficient, resulting in a high frequency of transcription initiation and effective elongation. These features result in an RNA elongation that is approximately five-fold faster than for E. coli RNA polymerase; hence, the T7 promoter is a much stronger promoter than the E. coli promoter.
T7 Promoter
5′ TAATACGACTCACTATAG 3′
T7 RNA polymerase is very selective and efficient, resulting both in a high frequency of transcription initiation and effective elongation. T7 RNA polymerase starts transcription at the underlined G in the promoter sequence. The polymerase then transcribes using the opposite strand as a template from 5’->3’. The first base in the transcript will be a G.
The DNA contacts made by T7 RNA polymerase have been mapped during binding and during the subsequent initiation of transcription. The RNA polymerase alone protects 19 bases in a region from -21 to -3. The reason for some literature suggesting additional bases after this G is because synthesis of the hexanucleotide mRNA (TAATACGACTCACTATAGGGAGA), expands the length of the sequence protected by the RNA polymerase to 32 bases and stabilizes the complex1.
1. Ikeda R. A. and Richardson C. C. Interactions of the RNA polymerase of bacteriophage T7 with its promoter during binding and initiation of transcription PNAS USA 83 (11): 3614-8 (1986).
No, the T7 RNA Polymerase recognizes the T7 Promoter sequence and then transcribes using the final G of the promoter as the first base to transcribe the opposite strand as a template from 5’->3’.
Higher yields of RNA may be obtained by raising rNTP concentrations and Mg2+ concentration (to 4 mM above the total NTP concentration) or increasing the plasmid DNA concentration (these would need to be titrated to determine the correct concentrations).
Additionally, inorganic pyrophosphatase can also be added to a final concentration of 4 units/mL to try to lower feedback inhibition caused by the build-up of pyrophosphates that bind and precipitate Mg2+ ions.
RNases are ubiquitous in the environment and a significant risk factor during RNA production. The activity of RNases is blocked using RNase inhibitor. We recommend using RNase inhibitor (MDX056) in the reaction.
Degraded the RNA product caused by RNase contamination is often the main cause. We recommend using RNase inhibitor (MDX056) in the reaction. The DNA should also be prepared free of NaCl, as T7 RNA Polymerase is sensitive to NaCl inhibition (concentration should not exceed 20 mM).
Get In Touch With A Specialist
Have questions about a product? Want to learn more about Meridian’s molecular or immunoassay reagent portfolio? We want to hear from you!