Air-Dryable™ qPCR and RT-qPCR Mixes Master Mixes
Meridians Air-Dryable™ mixes are glycerol-free mixes, ideal for developing ambient temperature, air-dried, multiplex qPCR and RT-qPCR molecular diagnostic tests and are suited for high-throughput, automated platforms or microfluidic qPCR platforms. Can also be used wet
Have questions about a product?
Meridian's Air-Dryable qPCR and RT-qPCR Master Mixes
- Create ultra-sensitive multiplex qPCR tests for low-copy number targets.
- Glycerol-free mixes are pre-formulated with a specialized blend of excipients, compatible with air-drying.
- High-tolerance to PCR inhibitors carried over during purification, improving reproducibility.
- Can be used wet or it can be dried, for developing an ambient-temperate stable assay.
Full enzyme activity following air-drying
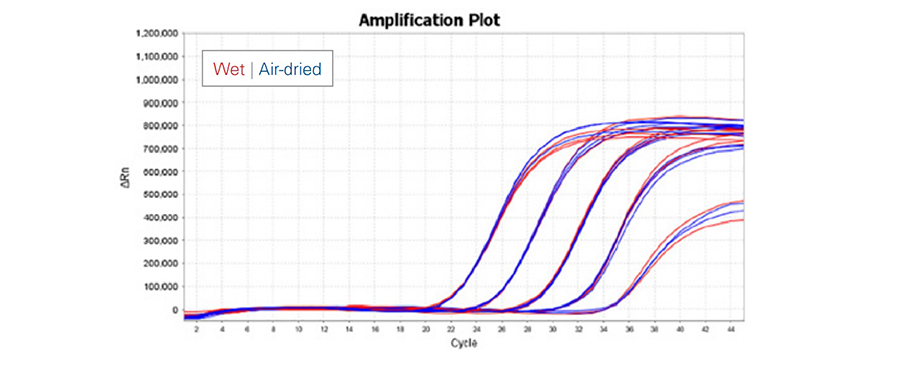
Activity of Air-Dryable™ 1-Step RT-qPCR Mix in both wet (red) and air-dried (blue) formats were compared on 10-fold dilution mouse RNA template. The air-dried mix showed no loss of activity and sensitivity when compared to freshly prepared wet mix up to the assay limit of detection.
Air-Dryable qPCR Mix, 4x, MDX082
Glycerol-free qPCR mix containing Taq polymerase, reaction buffer, dNTPs, MgCl2 and air-dry compatible excipients, ideal for developing ambient temperature, multiplex molecular assays.
Documents & Resources
Air-Dryable 1-Step RT-qPCR Mix, 4x, MDX095
Glycerol-free RT-qPCR mix containing Taq polymerase, reverse transcriptase, reaction buffer, dNTPs, MgCl2 and air-dry compatible excipients, ideal for developing ambient temperature, multiplex molecular assays.
Documents & Resources
Description
Molecular diagnostic tests are progressively moving towards dried formats. There are several advantages for this, including ambient temperature shipping and storage, extended shelf-life, reduced operating steps and potential errors and increased flexibility in sample volume. In order to be compatible with air or oven drying, however, enzyme preparations must be glycerol-free and include specialized excipients that preserve the mixture as it is exposed to higher temperatures.
Air-Dryable™ qPCR Mix (4x) is used for the development of multiplex molecular diagnostic tests, enabling highly sensitive detection of target DNA, whereas the Air-Dryable ™ 1-Step RT-qPCR Mix (4x) can be used for the development of multiplex molecular diagnostic tests both RNA and DNA targets. The air drying does not affect the specificity or reproducibility of the assay but increases the flexibility of the volume of sample that can be added, increasing sensitivity and also makes them ideal for microfluidic qPCR and point of care platforms.
Specifications
| Description | Glycerol-free qPCR and RT-qPCR mix containing Taq polymerase, reverse transcriptase, reaction buffer, dNTPs, MgCl2 and air-dry compatible excipients, ideal for developing ambient temperature, multiplex molecular assays. |
| Concentration | 4x |
| Appearance | Clear, colorless solution |
| Hot Start | Antibody mediated |
| Application | Probe-based, real-time PCR, two-step RT-qPCR, one-step RT-qPCR |
| Sample type | cDNA, crude or purified RNA and/or DNA |
| Presentation | 1 vial |
| Storage | -20 °C |
| Mix stability | See outer label |
| Assay stability | Up to 24 months at ambient temperature following air-drying |
| Consistency | ± 0.5 Ct variance between test and reference sample |
| DNase/RNase Contamination | No detectable degradation |
Catalogs & Brochures
FAQs: Air-Dryable qPCR and RT-qPCR Master Mixes
What causes low efficiency in qPCR?
Slope of standard 10-fold dilution curve indicates qPCR efficiency. The efficiency of the qPCR should be between 90–110% (−3.58 ≥ slope ≥ −3.1). If the efficiency is greater than 100%, this indicated inaccurate sample and reagent pipetting, if it is less than 100%, this indicates your samples may contain PCR inhibitors or your qPCR primer and/or probe design may not be optimal.
What are the advantages of using a one-step or a two-step?
One-step reaction
• Accurate representation of target copy number
• Simple and rapid
• Fewer pipetting steps (reducing possible errors and contamination)
• Best option for high-throughput screening
• Best method when only a few assays are run repeatedly
• Multiplex qPCR of gene of interest and control can be done in single well, from same RNA sample
Two-step reaction
• Two buffers optimized for independent RT and qPCR
• Highly sensitive
• Potentially more efficient because random primers and oligo d(T) can be used
• Possibility to stock cDNA to quantify several targets
• Recommended when the reaction is performed with a limiting amount of starting material
So one-step workflows are commonly favoured in molecular diagnostic tests. Two-step RT-qPCR is preferred when multiple interrogations will be made of the same starting material or where archiving of cDNA may be required.
Are reaction conditions the same for air-dried and liquid mixes?
Yes, reaction conditions are the same and they will produce the same Ct values, even with multiplex qPCR assays. This means that the liquid mixes can be used to create SOPs if you do not want to dry down and then later if drying is required (for example to increase sensitivity), the SOP can be updated for oven drying, it does not require completely new SOPs to be written.
What probes can be used in quantitative PCR?
Many fluorescent qPCR primer- and probe-based chemistries have been devised and are available from different commercial vendors for molecular diagnostic tests, the two most commonly used are:
• Hydrolysis (TaqMan) probes. The main advantages of using hydrolysis probes are high specificity, a high signal-to-noise ratio, and the ability to perform multiplex qPCR reactions. The disadvantages are that the initial cost of the probe.
• Molecular beacons. Molecular beacons are highly specific, can be used for multiplexing, and if the target sequence does not match the beacon sequence exactly, hybridization and fluorescence will not occur. Unlike hydrolysis probes, molecular beacons are displaced but not destroyed during amplification and can be used for melt curve analysis if necessary. The main disadvantage is that they are difficult to design, requiring a stable hairpin stem that is strong enough that the molecule will not spontaneously fold into non-hairpin conformations but not be too strong, or it may not properly hybridize to the target.
Can the Air-Dryable™ qPCR and RT-qPCR master mixes be used with crude samples?
The Air-Dryable™ qPCR and RT-qPCR master mixes have some inhibitor tolerance, but for amplification from crude lysates or inhibitor-rich samples we have the universal Inhibitor-Tolerant qPCR/RT-qPCR mixes and for even greater sensitivity, we have the air-dryable sample specific (blood, saliva, urine, stool, and plant) mixes.
Should I include a No-RT control reaction with my RT-qPCR?
No-RT control reactions are useful for determining issues that may arise from the amplification of genomic DNA that may be present in a sample. However, many RNA transcripts are present at low abundance and for these transcripts, it is recommended to perform a No-RT control reaction if primer sets do not span exon-exon junctions. If the primers do span exon-exon junctions, or genomic DNA is not likely to interfere (when looking at RNA viruses in a sample for example) a No-RT control is not required.
How long are the air-dried mixes stable for at ambient temperature?
Following air drying in the presence or absence of primers and probes, the Air-Dryable™ qPCR and RT-qPCR master mixes are stable for a minimum of 24 months at ambient temperature (17 – 23 °C). Following rehydration, the assay reproducibility, sensitivity and robustness will be the same as for a freshly made liquid mix, making the mixes ideal for point-of-care molecular diagnostic tests and microfluidic qPCR devices.
Get In Touch With A Specialist
Have questions about a product? Want to learn more about Meridian’s molecular or immunoassay reagent portfolio? We want to hear from you!
By submitting your information in this form, you agree that your personal information may be stored and processed in any country where we have facilities or service providers, and by using our “Contact Us” page you agree to the transfer of information to countries outside of your country of residence, including to the United States, which may provide for different data protection rules than in your country. The information you submit will be governed by our Privacy Statement.